Biology has fully embraced big-data, high-throughput approaches, and data workflows (Figure below) have become indispensable for processing, analyzing, understanding, and enabling new discoveries. These workflows begin with the interpretation of raw signals from laboratory instruments, such as DNA sequencers and mass spectrometers, and proceed through multiple steps of aggregation and filtering. This preprocessing stage transforms complex data into a biologically interpretable form, such as mutations statuses or levels of gene transcripts and proteins, and sets the stage for modeling and analysis. The data modeling and analysis stage uses various data science approaches to generate new insights into functioning of living organisms or finding drivers of disease that could potentially turn into new drugs, drug targets, or clinical biomarkers.
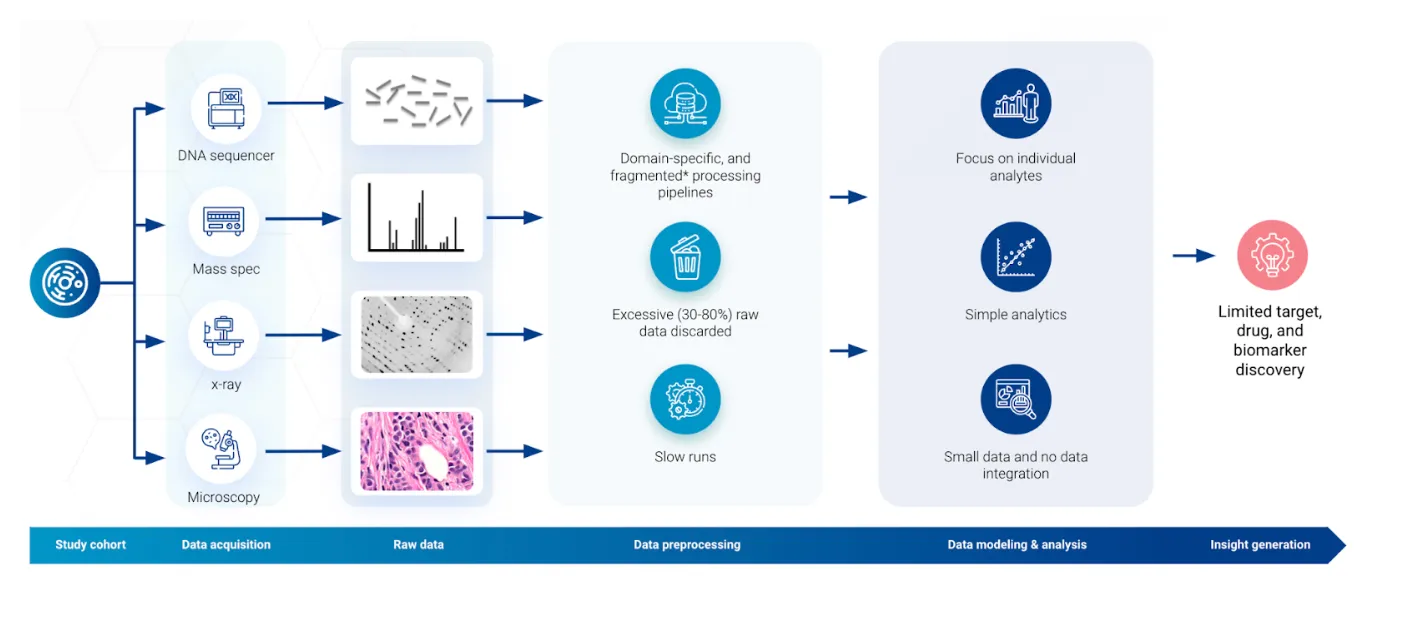
Yet, amidst the massive popularity of data science and ever-evolving data workflows, a significant imbalance has emerged between the two stages. The spotlight is usually on the latter stage of these workflows, where data analysis promises direct insights into biological functions and disease mechanisms. This focus is understandable, given the high impact of that one discovery capable of catalyzing major publications or spawning companies. However, this leaves the initial stage of raw data preprocessing in the shadows, often overlooked and even deemed as "unsexy," despite its critical importance.
Considering the high failure rate of novel discovery in life science research, the overlooked early stage of data processing presents a great opportunity for enhancing the success rate of life science research. Traditionally constrained by resources and legacy systems, preprocessing software is commonly designed to run on single workstations, often with limitations imposed by compute bandwidth and algorithms that are optimized for speed not accuracy. Current algorithms and approaches are not only slow and impose major bottlenecks in data generation workflows, but also unnecessarily simplify complex raw data, which can easily overlook and discard biologically significant findings, leading to substantial information loss even before the analysis stage begins.
We propose a reconsideration of these early, neglected steps in the data workflows. By acknowledging and addressing the limitations inherent in raw data preprocessing, we can unlock new potentials for life science research. The case of mass spectrometry proteomics is particularly illustrative, where up to 80% of measured analytes in a sample may be discarded due to data-loss issues. Recovering this "lost" information could dramatically influence drug discovery and disease treatment.
In our upcoming blog post, we will delve deeper into the challenges and opportunities within proteomics data processing. By shining a light on the early stages of data workflows, we aim to unveil strategies that can mitigate information loss and significantly enhance the impact of life science research. Stay tuned as we explore how revisiting the current data preprocessing could pave the way for the next wave of scientific breakthroughs.
Further read:
Recognizing millions of consistently unidentified spectra across hundreds of shotgun proteomics datasets [link]
Peptide splicing by the proteasome [link]
A universal SNP and small-indel variant caller using deep neural networks [link]: This research demonstrates how better algorithms can improve the accuracy of DNA sequencing. ()
Stay Ahead with Tesorai Insights
Receive the latest updates and industry insights from Tesorai. Topics of interest include machine learning, life sciences, and new Tesorai releases.
